K-Means Clustering with Python and Spark
Imagine that you have a customer dataset, and you need to apply customer segmentation on this historical data.
Customer segmentation is the practice of partitioning a customer base into groups of individuals that have similar characteristics.
It is a significant strategy as a business can target these specific groups of customers and effectively allocate marketing resources.
For example, one group might contain customers who are high-profit and low-risk, that is, more likely to purchase products, or subscribe for a service.
Table of contents
The installation of Python and Pyspark and the introduction of K-Means is given here.
K-Means Clustering with Python
import random
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
%matplotlib inline
import pandas as pd
cust_df = pd.read_csv("Cust_Segmentation.csv")
cust_df.head()
| Customer Id | Age | Edu | Years Employed | Income | Card Debt | Other Debt | Defaulted | Address | DebtIncomeRatio | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 41 | 2 | 6 | 19 | 0.124 | 1.073 | 0.0 | NBA001 | 6.3 |
| 1 | 2 | 47 | 1 | 26 | 100 | 4.582 | 8.218 | 0.0 | NBA021 | 12.8 |
| 2 | 3 | 33 | 2 | 10 | 57 | 6.111 | 5.802 | 1.0 | NBA013 | 20.9 |
| 3 | 4 | 29 | 2 | 4 | 19 | 0.681 | 0.516 | 0.0 | NBA009 | 6.3 |
| 4 | 5 | 47 | 1 | 31 | 253 | 9.308 | 8.908 | 0.0 | NBA008 | 7.2 |
df = cust_df.drop('Address', axis=1)
df.head()
| Customer Id | Age | Edu | Years Employed | Income | Card Debt | Other Debt | Defaulted | DebtIncomeRatio | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 41 | 2 | 6 | 19 | 0.124 | 1.073 | 0.0 | 6.3 |
| 1 | 2 | 47 | 1 | 26 | 100 | 4.582 | 8.218 | 0.0 | 12.8 |
| 2 | 3 | 33 | 2 | 10 | 57 | 6.111 | 5.802 | 1.0 | 20.9 |
| 3 | 4 | 29 | 2 | 4 | 19 | 0.681 | 0.516 | 0.0 | 6.3 |
| 4 | 5 | 47 | 1 | 31 | 253 | 9.308 | 8.908 | 0.0 | 7.2 |
Normalizing over the standard deviation
Now let’s normalize the dataset. But why do we need normalization in the first place? Normalization is a statistical method that helps mathematical-based algorithms to interpret features with different magnitudes and distributions equally. We use StandardScaler() to normalize our dataset.
from sklearn.preprocessing import StandardScaler
X = df.values[:,1:]
X = np.nan_to_num(X)
Clus_dataSet = StandardScaler().fit_transform(X)
Clus_dataSet
array([[ 0.74291541, 0.31212243, -0.37878978, ..., -0.59048916,
-0.52379654, -0.57652509],
[ 1.48949049, -0.76634938, 2.5737211 , ..., 1.51296181,
-0.52379654, 0.39138677],
[-0.25251804, 0.31212243, 0.2117124 , ..., 0.80170393,
1.90913822, 1.59755385],
...,
[-1.24795149, 2.46906604, -1.26454304, ..., 0.03863257,
1.90913822, 3.45892281],
[-0.37694723, -0.76634938, 0.50696349, ..., -0.70147601,
-0.52379654, -1.08281745],
[ 2.1116364 , -0.76634938, 1.09746566, ..., 0.16463355,
-0.52379654, -0.2340332 ]])
clusterNum = 3
k_means = KMeans(init = "k-means++", n_clusters = clusterNum, n_init = 12)
k_means.fit(X)
labels = k_means.labels_
#print(labels)
We assign the labels to each row in dataframe.
df["Clus_km"] = labels
df.head(5)
| Customer Id | Age | Edu | Years Employed | Income | Card Debt | Other Debt | Defaulted | DebtIncomeRatio | Clus_km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 41 | 2 | 6 | 19 | 0.124 | 1.073 | 0.0 | 6.3 | 0 |
| 1 | 2 | 47 | 1 | 26 | 100 | 4.582 | 8.218 | 0.0 | 12.8 | 2 |
| 2 | 3 | 33 | 2 | 10 | 57 | 6.111 | 5.802 | 1.0 | 20.9 | 0 |
| 3 | 4 | 29 | 2 | 4 | 19 | 0.681 | 0.516 | 0.0 | 6.3 | 0 |
| 4 | 5 | 47 | 1 | 31 | 253 | 9.308 | 8.908 | 0.0 | 7.2 | 1 |
We can easily check the centroid values by averaging the features in each cluster.
df.groupby('Clus_km').mean()
| Customer Id | Age | Edu | Years Employed | Income | Card Debt | Other Debt | Defaulted | DebtIncomeRatio | |
|---|---|---|---|---|---|---|---|---|---|
| Clus_km | |||||||||
| 0 | 432.468413 | 32.964561 | 1.614792 | 6.374422 | 31.164869 | 1.032541 | 2.104133 | 0.285185 | 10.094761 |
| 1 | 410.166667 | 45.388889 | 2.666667 | 19.555556 | 227.166667 | 5.678444 | 10.907167 | 0.285714 | 7.322222 |
| 2 | 402.295082 | 41.333333 | 1.956284 | 15.256831 | 83.928962 | 3.103639 | 5.765279 | 0.171233 | 10.724590 |
Now, lets look at the distribution of customers based on their age and income:
area = np.pi * ( X[:, 1])**2
plt.scatter(X[:, 0], X[:, 3], s=area, c=labels.astype(np.float), alpha=0.5)
plt.xlabel('Age', fontsize=18)
plt.ylabel('Income', fontsize=16)
plt.show()
C:\Anaconda3\envs\pyspark\lib\site-packages\ipykernel_launcher.py:2: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
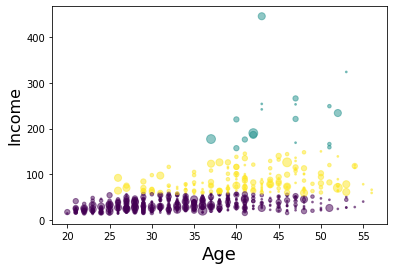
k-means will partition your customers into mutually exclusive groups, for example, into 3 clusters. The customers in each cluster are similar to each other demographically.
K-Means Clustering with Pyspark
First thing to do is start a Spark Session
import findspark
findspark.init()
from pyspark.sql import SparkSession
spark = SparkSession.builder.appName('customers').getOrCreate()
from pyspark.ml.clustering import KMeans
# Loads data.
dataset = spark.read.csv("Cust_Segmentation.csv",header=True,inferSchema=True)
dataset.head(1)
[Row(Customer Id=1, Age=41, Edu=2, Years Employed=6, Income=19, Card Debt=0.124, Other Debt=1.073, Defaulted=0, Address='NBA001', DebtIncomeRatio=6.3)]
#dataset.describe().show(1)
dataset.printSchema()
root
|-- Customer Id: integer (nullable = true)
|-- Age: integer (nullable = true)
|-- Edu: integer (nullable = true)
|-- Years Employed: integer (nullable = true)
|-- Income: integer (nullable = true)
|-- Card Debt: double (nullable = true)
|-- Other Debt: double (nullable = true)
|-- Defaulted: integer (nullable = true)
|-- Address: string (nullable = true)
|-- DebtIncomeRatio: double (nullable = true)
As you can see, Address in this dataset is a categorical variable. k-means algorithm isn’t directly applicable to categorical variables because Euclidean distance function isn’t really meaningful for discrete variables. So, lets drop this feature and run clustering.
columns_to_drop = ['Address']
dataset = dataset.drop(*columns_to_drop)
dataset.printSchema()
root
|-- Customer Id: integer (nullable = true)
|-- Age: integer (nullable = true)
|-- Edu: integer (nullable = true)
|-- Years Employed: integer (nullable = true)
|-- Income: integer (nullable = true)
|-- Card Debt: double (nullable = true)
|-- Other Debt: double (nullable = true)
|-- Defaulted: integer (nullable = true)
|-- DebtIncomeRatio: double (nullable = true)
dataset.columns
['Customer Id',
'Age',
'Edu',
'Years Employed',
'Income',
'Card Debt',
'Other Debt',
'Defaulted',
'DebtIncomeRatio']
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
from pyspark.sql.types import IntegerType
dataset = dataset.withColumn("Defaulted", dataset["Defaulted"].cast(IntegerType()))
dataset.printSchema()
root
|-- Customer Id: integer (nullable = true)
|-- Age: integer (nullable = true)
|-- Edu: integer (nullable = true)
|-- Years Employed: integer (nullable = true)
|-- Income: integer (nullable = true)
|-- Card Debt: double (nullable = true)
|-- Other Debt: double (nullable = true)
|-- Defaulted: integer (nullable = true)
|-- DebtIncomeRatio: double (nullable = true)
feat_cols = [
'Age',
'Edu','Years Employed','Income','Card Debt','Other Debt','DebtIncomeRatio']
vec_assembler = VectorAssembler(inputCols = feat_cols, outputCol='features')
final_data = vec_assembler.transform(dataset)
from pyspark.ml.feature import StandardScaler
scaler = StandardScaler(inputCol="features", outputCol="scaledFeatures", withStd=True, withMean=False)
final_data
DataFrame[Customer Id: int, Age: int, Edu: int, Years Employed: int, Income: int, Card Debt: double, Other Debt: double, Defaulted: int, DebtIncomeRatio: double, features: vector]
# Compute summary statistics by fitting the StandardScaler
scalerModel = scaler.fit(final_data)
# Normalize each feature to have unit standard deviation.
cluster_final_data = scalerModel.transform(final_data)
Train the Model and Evaluate
** Time to find out whether its 2 or 3! **
# Trains a k-means model.
kmeans3 = KMeans(featuresCol='scaledFeatures',k=3)
kmeans2 = KMeans(featuresCol='scaledFeatures',k=2)
model3 = kmeans3.fit(cluster_final_data)
model2 = kmeans2.fit(cluster_final_data)
from pyspark.ml.clustering import KMeans
from pyspark.ml.evaluation import ClusteringEvaluator
# Make predictions
predictions3 = model3.transform(cluster_final_data)
predictions2 = model2.transform(cluster_final_data)
# Evaluate clustering by computing Silhouette score
evaluator = ClusteringEvaluator()
silhouette = evaluator.evaluate(predictions3)
print("With k=3 Silhouette with squared euclidean distance = " + str(silhouette))
silhouette = evaluator.evaluate(predictions2)
print("With k=2 Silhouette with squared euclidean distance = " + str(silhouette))
With k=3 Silhouette with squared euclidean distance = 0.05288868077858952
With k=2 Silhouette with squared euclidean distance = 0.6581986063262965
#Show the results
centers=model.clusterCenters()
print("Cluster Centers:")
for center in centers:
print(center)
Cluster Centers:
[5.38544237 2.08381863 2.54750121 2.6356673 1.9964595 2.30786829
2.04421758]
[3.71924395 2.29195271 0.55736835 0.78361889 0.68066746 0.87084406
2.18192288]
[4.38305208 1.49522293 1.24517269 0.97828653 0.34932727 0.44759616
0.93527274]
# Evaluate clustering by computing Within Set Sum of Squared Errors.
for k in range(2,9):
kmeans = KMeans(featuresCol='scaledFeatures',k=k)
model = kmeans.fit(cluster_final_data)
predictions = model.transform(cluster_final_data)
evaluator = ClusteringEvaluator()
silhouette = evaluator.evaluate(predictions)
print("With K={}".format(k))
print("Silhouette with squared euclidean distance = " + str(silhouette))
print('--'*30)
With K=2
Silhouette with squared euclidean distance = 0.6581986063262965
------------------------------------------------------------
With K=3
Silhouette with squared euclidean distance = 0.05288868077858952
------------------------------------------------------------
With K=4
Silhouette with squared euclidean distance = 0.23934815694787387
------------------------------------------------------------
With K=5
Silhouette with squared euclidean distance = 0.09503424227629984
------------------------------------------------------------
With K=6
Silhouette with squared euclidean distance = 0.03747870420927804
------------------------------------------------------------
With K=7
Silhouette with squared euclidean distance = 0.07544170107415611
------------------------------------------------------------
With K=8
Silhouette with squared euclidean distance = 0.07633841797382555
------------------------------------------------------------
Let’s check with the transform and prediction columns that result form this! Congratulations if you made this connection, it was quite tricky given what we’ve covered!
model3.transform(cluster_final_data).groupBy('prediction').count().show()
+----------+-----+
|prediction|count|
+----------+-----+
| 1| 261|
| 2| 439|
| 0| 150|
+----------+-----+
model2.transform(cluster_final_data).groupBy('prediction').count().show()
+----------+-----+
|prediction|count|
+----------+-----+
| 1| 183|
| 0| 667|
+----------+-----+
You can download the notebook here
Congratulations! We have practiced K-Means Clustering with Python and Spark.

Leave a comment